Apps
Soft-matter-based Nanoscale Systems
Cancer
COVID-19
General Multicellular Systems
Basic Biophysics and Training
Externally Developed Apps
Soft-matter-based Nanoscale Systems
Ionic Self Assembly
 Ions in Nanoconfinement (nano)
Ions in Nanoconfinement (nano)
This app empowers users to simulate ions confined between material surfaces that are nanometers apart, and extract the associated ionic structure. The app facilitates investigations for a wide array of ionic and environmental parameters using standard molecular dynamics (MD) method for unpolarizable surfaces.
 Nanosphere Electrostatics Lab (nano)
Nanosphere Electrostatics Lab (nano)
This app allows users to simulate the self-assembly of ions near a spherically shaped nanoparticle, and extract the effective electrostatic properties..
Nanoparticle Self Assembly
 Nanoparticle Assembly Lab (nano)
Nanoparticle Assembly Lab (nano)
Simulate assembly of nanoparticles into aggregates in physiological conditions.
 Polyvalent Nanoparticle Binding Simulator (nano)
Polyvalent Nanoparticle Binding Simulator (nano)
Simulates the binding of ligand-decorated nanoparticles to cell membrane driven by ligand-cell-receptor attraction
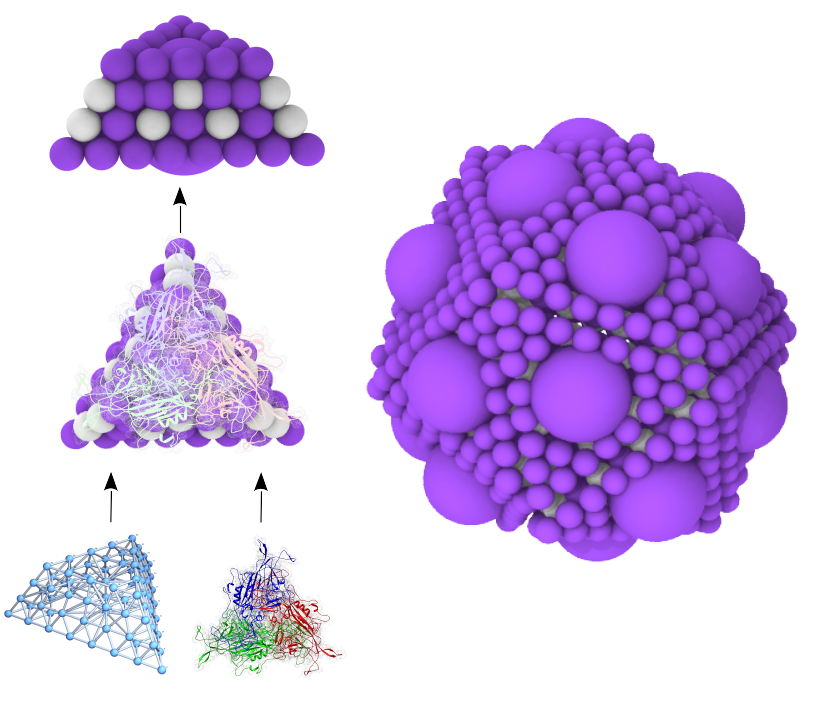 Souffle: Virus Capsid Assembly Lab (nano)
Souffle: Virus Capsid Assembly Lab (nano)
Simulate assembly of flexible capsomeres into virus capsids and oligomers.
 Nanoparticle Shape Lab (nano)
Nanoparticle Shape Lab (nano)
The shape of nanocontainers determines their ability to act as targeted drug delivery carriers by modulating their biodistribution and bioavailability. This app simulates the shape deformation of soft-matter-based deformable, charged nanocontainers for a broad variety of nanocontainer material properties and solution conditions. Molecular dynamics based simulated annealing is used to minimize the energy of the nanocontainer for a given set of material and solution parameters.
Cancer apps
 Compucell3D Vascular Tumor (micro)
Compucell3D Vascular Tumor (micro)
Simulate 3D vascular tumor with CompuCell3D
 PhysiCell: cancer nanotherapy simulator (macro)
PhysiCell: cancer nanotherapy simulator (macro)
This app allows users to design and explore new and novel nanotherapies, where engineered nanoparticles are conjugated to novel cancer therapeutics. Users can simulate the growth and migration of cancer cells in the tissue environment, as well as pharamcodynamic response to user-designed therapeutics. Adapted from the PhysiCell Framework
 PhysiCell: cancer biorobots simulation (macro)
PhysiCell: cancer biorobots simulation (macro)
2D simulation of biorobots delivering a drug to cancer cells
 PhysiCell: cancer-immune model (macro)
PhysiCell: cancer-immune model (macro)
PhysiCell model of immune cells attacking a heterogeneous tumor
 PhysiCell: tumor heterogeneity (macro)
PhysiCell: tumor heterogeneity (macro)
2D simulation of tumor heterogeneity
 PhysiCell: liver tissue mechanobiology (macro)
PhysiCell: liver tissue mechanobiology (macro)
Simulates the liver tissue mechanobiology on cancer metastatic seeding.
 PhysiCell: AMIGOS - Hypoxia (macro)
PhysiCell: AMIGOS - Hypoxia (macro)
Hypoxic conditions occur in the center of tumor after tumor cells use and deplete oxygen. Hypoxic cells secrete signaling chemicals to induce vascularization, such as vascular epithelial growth factor (VEGF).
CompuCell3D - Simulation of Angiogenesis
In this tool we simulate a bacterium secreting a diffusive chemical signal, the macrophage is able to detect the gradient of this signal and thus hunt the macrophase. In CompuCell3D the default view is of the cells, we can change the view to observe the chemical field by selecting the drop-down menu in the "Graphics 0" window and selecting it (in the case of this tool the chemical is named ATTR). If you don't see the cells anymore when changing to this view make sure that cell borders is selected under Visualizations option menu.
PhysiCell Model for Tumor Hypoxia
This app demonstrates the evaluation of the new hypotheses on the frequency and duration of phenotypic changes in cancer cells under the influence of hypoxic conditions.
This model and cloud-hosted demo are part of a collaboration between Dr. Macklin's lab and Dr. Gilkes's lab, through of a project granted by Jayne Koskinas Ted Giovanis (JKTG) Foundation for Health and Policy and the Breast Cancer Research Foundation (BCRF). It is also part of the education and outreach for the IU Engineered nanoBIO Node and the NCI-funded cancer systems biology grant U01CA232137. The model is built using PhysiCell: a C++ framework for multicellular systems biology [1].
COVID-19 apps
 COVID 19 Virtual Tissue Model - Tissue Infection and Immune Response Dynamics (micro)
COVID 19 Virtual Tissue Model - Tissue Infection and Immune Response Dynamics (micro)
Simulates tissue and immune system interactions during a viral lung infection
 PhysiCell for COVID-19 (macro)
PhysiCell for COVID-19 (macro)
This model simulates replication dynamics of SARS-CoV-2 (coronavirus / COVID19) in a layer of epithelium. It is being rapidly prototyped and refined with community support. See related: PC4COVID19: Rapid Community Prototyping of a Coronavirus (COVID-19) Replication Simulator
COVID-19 Data Analysis Educational Tool
This educational tool provides users with a Jupyter notebook that teaches them how to use Python to analyze and visualize some pre-computed data from the https://nanohub.org/tools/pc4covid19 app. Because it is an interactive notebook, users can edit the Python code and explore on their own.
PhysiCell: Infected System and Off-Site Immune System
In this App, we developed a dynamic interaction model which contains two systems: an infected system of cells and a off-site immune system that respond to the infection. We observe that immune response changes depending on the intensity of the infection cells. Our aim is to investigate how the immune system modulate its response according to variation in the infection.
General Multicellular Systems
 CompuCellSort (micro)
CompuCellSort (micro)
simulate cell sorting with compucell 3d
 PhysiCell: Thanos vs. Avengers multicellular demonstrator (macro)
PhysiCell: Thanos vs. Avengers multicellular demonstrator (macro)
This app demonstrates chemotactic migration, cell-cell contact interactions, and iterating across cells to apply a random event. It also explores the impact of varying the number and strength of different cells in a complex, stochastic adversarial system.
CompuCell3D v4 Main Tool
CompuCell3D is a flexible scriptable modeling environment, which allows the rapid construction of sharable Virtual Tissue in-silico simulations of a wide variety of multi-scale, multi-cellular problems including angiogenesis, bacterial colonies, cancer, developmental biology, evolution, the immune system, tissue engineering, toxicology and even non-cellular soft materials. CompuCell3D models have been used to solve basic biological problems, to develop medical therapies, to assess modes of action of toxicants and to design engineered tissues. CompuCell3D intuitive and make Virtual Tissue modeling accessible to users without extensive software development or programming experience. It uses Cellular Potts Model to model cell behavior.
CompuCell3D v3
An open-source simulation environment for multi-cell, single-cell-based modeling of tissues, organs and organisms. Uses Cellular Potts Model to model cell behavior. This version is still supported for models that are specifically developed for this version.
CompuCell3D 3D Cell Sorting
CompuCell3D based application to simulate 3D cell sorting
CompuCell3d cell sorting in a hexagonal lattice
Showcases hexagonal lattice use in CompuCell3D by simulating cell sorting by difference in contact energies in a hexagonal lattice.
CompuCell3D - 2D wet foam coarsening
CompuCell3D Demo for a 2D foam coarsening without drainage (gravity). This simulations seeds space with cells representing air bubbles, they are allowed to relax to their target volume with no other cell type present. On time step 200 the simulation's empty space is filled with cells representing water. The whole system is allowed to relax for another 100 time steps then air bubbles have their volume constrain removed, i.e. can freely change their volumes.
CompuCell3D - 2D wet foam coarsening with drainage
CompuCell3D Demo for a 2D foam coarsening with drainage (gravity). This simulations seeds space with cells representing air bubbles, they are allowed to relax to their target volume with no other cell type present. On time step 200 the simulation's empty space is filled with cells representing water. The whole system is allowed to relax for another 100 time steps then air bubbles have their volume constrain removed, i.e. can freely change their volumes. On time step 500 gravity (drainage) is turned on and water cells accumulate at the bottom of the simulation.
CompuCell3D - Delta-Notch signaling in a group of cells
CompuCell3D can solve individual cell's ODE and have the information of one cell affect another (implemented trough SBML). Cells participating in Delta-Notch signaling. You can visualize Delta and Notch levels field by selecting the drop-down menu in the "Graphics" window and selecting one of them (changing the chemical visualized pauses the simulation, you'll need to hit run after the change). You can have more than one "Graphics" window open by selecting the menu "Visualization->New Graphics Window"
PhysiCell biorobots simulation
2D simulation of director, worker, and cargo cells.
Three-Type Multicellular Simulation Lab
Inspired by the 3-body problem in physics, this virtual cell laboratory explores a tunable multicellular system with 3 interacting cell types. Similarly to the 3-body problem, simple cell-cell interactions can lead to complex emergent dynamics, including cooperativity, competition, oscillations, and chaotic phenomena.
This virtual cell lab lets you broadly explore the effects of your assumptions of how cells affect one another to drive the emergent multicelular dynamics.
Basic Biophysics and Training
 Compucell3D Bacterium Macrophage (micro)
Compucell3D Bacterium Macrophage (micro)
Simulation of macrophage chasing bacterium
 PhysiCell: biased random migration generator (macro)
PhysiCell: biased random migration generator (macro)
PhysiCell demonstration of biased random cell migration
PhysiCell: Motility Training App (macro)
To account for cell movement, PhysiCell includes a set of parameters that govern speed and direction of cell motility. The biased, random motility of cells is determined by the following equation:where b is the level of bias in movement (migration bias), ξ is a random unit vector of length 1, and dbias is the direction.
 PhysiCell: invader-scout-attacker system (macro)
PhysiCell: invader-scout-attacker system (macro)
PhysiCell model of an invader-scout-attacker multicellular system
 PhysiCell workshop training (macro)
PhysiCell workshop training (macro)
This tool is intended to be used at PhysiCell training workshops. It is general purpose in the sense that it provides a GUI that includes 1) a Terminal tab to compile and run a PhysiCell model, and 2) a Plotting tab to visualize results.
Volume Training App for PhysiCell
Training application for "Volume" concept in PhysiCell.
Motility Training App
Training application for "Motility" concept in PhysiCell. To account for cell movement, PhysiCell includes a set of parameters that govern speed and direction of cell motility. The biased, random motility of cells is determined by the following equation:
where b is the level of bias in movement (migration bias), ξ is a random unit vector of length 1, and dbias is the direction of the bias.
Death Training App for PhysiCell
Training application for "Death" concept in PhysiCell. Cells do not keep growing and dividing forever. Eventually, they undergo cell death, in which cells lyse (burst) and decay. PhysiCell supports two models of cell death: apoptosis and necrosis. Apoptosis is programmed cell death, which contributes to regular growth and function. Necrosis, on the other hand, is unprogrammed cell death due to injury or disease. PhysiCell users may choose the model(s) appropriate for their projects.
Microenvironment Concept Training App for PhysiCell
Training application for "Microenvironment" concept in PhysiCell.
Mechanics Training App for PhysiCell
Training application for "Mechanics" concept in PhysiCell.
Externally Developed Apps
 PhysiCell: Replication Competent Oncolytic Virus in Epithelial cell layer (macro)
PhysiCell: Replication Competent Oncolytic Virus in Epithelial cell layer (macro)
This app demonstrates the interaction between a replication competent oncolytic viruse and a layer of epithelial cells. In this model, epithelial cells (pink) divide while consuming oxygen, with greater availability of oxygen driving faster proliferation.
 PhysiCell: Death Training App
PhysiCell: Death Training App
Cells do not keep growing and dividing forever. Eventually, they undergo cell death, in which cells lyse (burst)and decay. PhysiCell supports two models of cell death: apoptosis and necrosis. Apoptosis is programmed cell death, which contributes to regular growth and function.
 PhysiCell: Extracellular Matrix Modeling (macro)
PhysiCell: Extracellular Matrix Modeling (macro)
An extracellular matrix model developed in PhysiCell
 PhysiCell: Wound Healing
PhysiCell: Wound Healing
In silico demonstration of in vitro scratch wound healing using PhysiCell
 PhysiBoSS Simulation with ECM
PhysiBoSS Simulation with ECM
Agent based multicellular simulation with extracellular matrix interaction.
PhysiCell: infected system and off-site immune system
In this app, we developed a dynamic interaction model which contains two systems: an infected system of cells and a off-site immune system that responds to the infection.
Replication Competent Oncolytic Virus expressing secretable trimeric TRAIL: hypothesis testing
This app demonstrates the interaction between a replication competent oncolytic viruse and a layer of epithelial cells. It uses the PhysiCell software published PhysiCell in Ghaffarizadeh et al. (2018). In this model, epithelial cells (pink) divide while consuming oxygen, with greater availability of oxygen driving faster proliferation.
PhysiBoSS: cell fate decision in TNF Boolean model
This app demonstrates the population dynamics in a multicellular system where cells run a Boolean network model representing the TNF signalling pathway and when exposed to TNF.
Spatial Pattern Formation in Cell Based Models
Multi-species Biofilm on Wound Surface
This tool aims to model two bacterial species which form a biofilm on the surface of a chronic wound.
Replication Competent Oncolytic Virus in Epithelial cell layer - Adrianne Jenner
PhysiCell simulation of a replication competent oncolytic virus release from two veins, diffusing through a layer of epithelial cells.
PhysiBoSS simulation of COVID19 infection - Institut Curie and BSC
PhysiBoSS-COVID: the Boolean modelling of COVID-19 signalling pathways in a multicellular simulation framework
Others
